What is a bio-computer? What does biological computing power mean in computers?
While meteorological institutes produce global weather claims, many different information flows to these assumptions throughout the process …

While meteorological institutes produce global weather claims, many different information flows to these assumptions throughout the process. Weather researchers use computers with many different processors to better cope with the information metric. Each of these processors, called “parallel computers”, simultaneously costs data from a reasonable part of the world.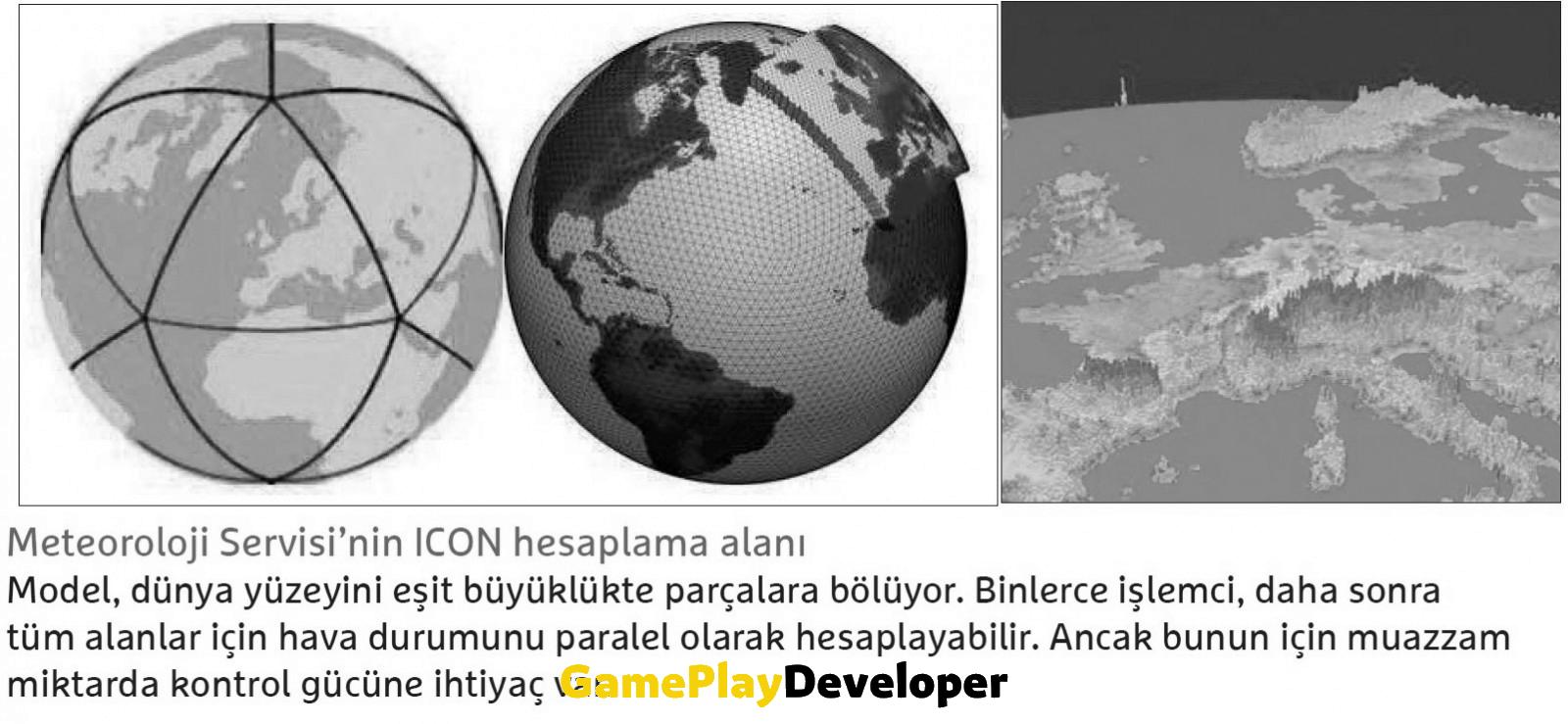
In addition to the arithmetic units that perform the actual processes, such a computer also needs many processors just to distribute the processes. Simply put, they assign missions to arithmetic units and combine partial results into a logical aggregate result. In the example of the weather claim, the controlling processors individually control the calculations from the sub-areas of the Earth. They organize the exchange of information and results in their midst. When we look at it, it is useless to assume how a high pressure area will behave if the influence of low pressure areas in the immediate vicinity is not taken into account.
Computing Power for the Control System
However, you can’t simply connect an arbitrary number of processors together to solve an arbitrary number of complex problems. The more computers running in parallel, the more information processing power is required for the control system. The effort required for control ambitions often increases disproportionately. This means that there are only a few applications where parallel computing seems to be a viable option.
The reason lies in the architecture of a digital computer. It was designed to run one information process step after another. In recent years, the increasing computing power of computers has enabled digital computers to solve increasingly complex problems despite this hassle. However, the further development of digital computers always faces physical and technical limits. Thus, to make parallel computation easier, other research approaches to computation are of interest.
wins.
In the shadow of quantum computers, which are attracting great attention with their promising research results in this field, many teams It also takes steps with approaches in the field of biology.
What is a Bio-Computer?
The international research project Bio-4Comp has been dealing with networked biocomputers for several years. The idea is that biological spies are sent on a journey through a complex network of nanochannels that represents a mathematical problem. A number is then added or not added to the cumulative result, depending on which turn the spy makes at an intersection. The path the agent takes over the network corresponds to a possible outcome.
The advantage is that you can send many agents over the network at the same time, and they can monitor all potential routes one-on-one. Therefore, instead of properly calculating one assay path after another like a conventional digital computer, the network-based biocomputer needs to do the calculations in parallel. of the Fraunhofer Institute for Electronic Nano Systems (ENAS) in Chemnitz. Danny Reuter is responsible for research work on the fabrication of networks and the scalability of technologies.
Dr. Reuter compares crowd surfing at a rock concert to the midst of processes in the network: “Motor proteins move biological spies, which in our case are molecules derived in animal cells, just as music fans carry a musician through an audience.” So here the team is moving spies over the net. It uses the kinetic power of motor proteins to
Thomas Blaudeck, Reuter’s colleague, again from Fraunhofer ENAS, hopes to have millions of agents in a network in the future to move from basic research to applied research: “Each spy is its own processor. Since moving around the nanonetwork is much slower than the information processing speed of a classical digital computer. , we need a large number of intermediaries to take advantage of our advantages in practical applications.“
Viruses as Processors
These advantages are primarily related to parallelism and power efficiency. These are exactly the areas where digital computers face challenges. Blaudeck sees potential applications of the network-based biocomputer, in principle, in all tasks of exponentially increasing complexity of possible combinations in each choice. “The advantage we have with biological approaches is the material. Because it can reproduce itself under reasonable conditions.” says. In Bio4Comp, groups work with dead elements that do not have a life of their own.
However, molecules acting in the network as spies can split, for example, at intersections, thus performing two computational steps simultaneously. The first part adds the number represented by the intersection, the second part takes a different path and does not add the number.
However, other research projects mainly work with live spies and send viruses or bacteria through networks. Here, middlemen can simply multiply to increase the number of processors. This duplication is first and foremost the most necessary action. Because a kind of “bottleneck” is created at the entrance of the network. There, only a finite number of agents can enter the network at any given time. But the network branches further and grows larger with each pass.
Networks that truly allow for practical computations need a large number of intersections to represent a complex problem. Blaudeck explains, “Agent density, that is, the number of spies coming from one channel portion per unit time, gets smaller and smaller as the output becomes real. Then biology helps us with this problem.” he explains.
A Complement for Supercomputers
One day, bio-computers may also excel in power efficiency. According to Danny Reuter, these computers cannot replace personal computers that sit under more than one person’s desk. “Our computers were designed to complement great computers. All the problems we want to solve with biocomputers can also be solved by great computers. But one day we hope to be faster and use a lot less power to do one-to-one calculations.” According to the two Fraunhofer researchers, three to four orders of magnitude—less energy per computation—is the goal of the projects.
However, there are still a few hurdles to overcome along the way. “So far we have been able to show that the approach works to a robust degree,” Reuter said. Right now our results are on the side of where quantum computers were three or four years ago and still far from competing with superior computers.” says. The crux of the annoyance is scaling, which is the main focus of Reuter and Blaudeck’s Fraunhofer team. “As we continue to grow our networks and send more agents, the space we need for a related issue has become enormous,” Reuter said. The defect rate would also be very high.” he states. This is where the next construction site is seen.
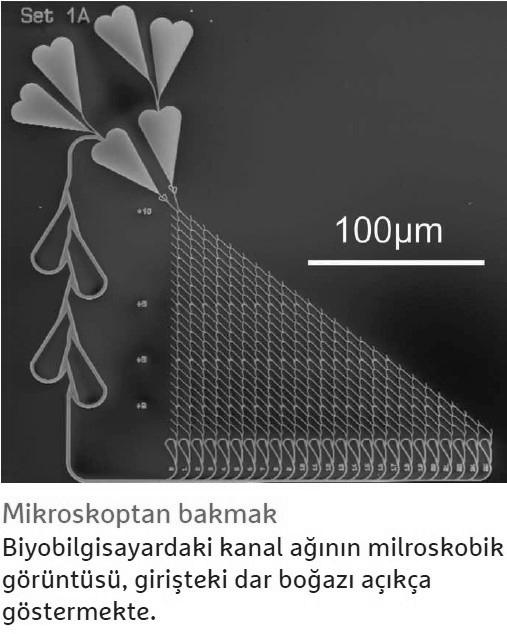
For example, tagging in words should improve the performance of computers. In this process, researchers mark molecules as they pass through the network so they can better read where they’re going later. Currently, Reuter reports that the spy is still viewed with a microscope as it travels through the network. “But we are working on electronic components that receive a signal when the spy passes by, or add a measure of DNA to it at a certain point in the network, and then track which path it takes.”
This will also facilitate detection at the exit of the network, which will be automated in the next step.
Not Yet Particularly Sustainable
Also interchangeable junctions are missing in the project. So far, a nanonetwork represents only a single math problem. Thomas Blaudeck explains that the networked computer blurs the end between hardware and software: “In our case, software is represented in hardware by the exact arrangement of junctions.” Another chip for each computation, something researchers agree, is now particularly unsustainable. However, various computational problems can be represented and computed with a single chip if universally interchangeable intersections can be implemented.
While many questions remain unresolved, Reuter and Blaudeck are in an optimistic frame of mind. The biotechnology and manufacturing technologies required to produce nanochannels already exist. The challenge here is both to bring together scientific disciplines with mathematics and computer science, and to develop sub-elements unfamiliar to classical microelectronics.
DNA to Calculate Square Roots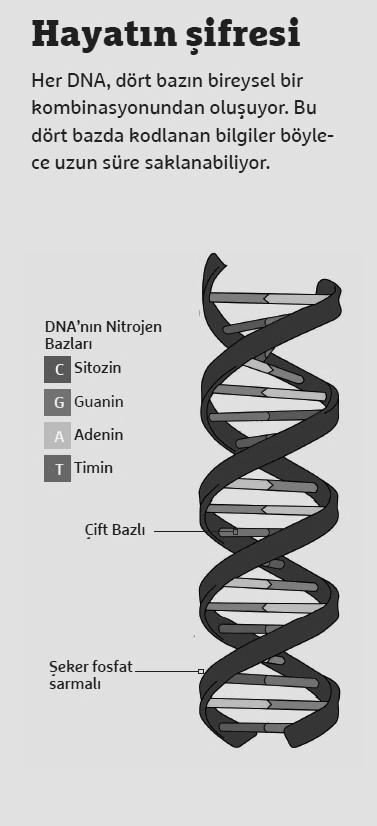
In the middle, research groups are tapping into other applications in biology. Computer scientist and molecular biologist Leonard Adleman conducted experiments with a programmable DNA in 1994. He then represented the input values in the DNA sequences that they reacted with each other in a test tube. With this, Adleman was able to perform easy mathematical calculations.
In 2019, another group was able to calculate square roots with such a DNA computer. Each DNA strand was given its own fluorescent color expense. The new combinations of these color values after the experiment then corresponded to the result of the calculation.
The advantage of this approach is massive parallelization, as with the networked biocomputer. DNA strands react with each other at the same time in all combinations in the test tube.
In theory, it’s particularly well suited for optimization problems. There are always several viable solutions to these problems. But one of them is the most sufficient, fastest and most economical. The most well-known example is the traveling salesman problem. A merchant should cover all the cities in a list without visiting any of them twice. There are countless travel note options that come before him, but he naturally wants to use the shortest route to save kilometers.
Evaluation Methods Not Available
In the DNA computer, each city would receive its own DNA strand. They would all react to each other by duplicating a “nuthouse”, thus creating all conceivable paths at the same time. It would take years for a digital computer to do this calculation for a certain number of cities. If you now remove longer DNA modules with targeted chemical responses, theoretically the shorter path remains of all.
The trick is this: Practically, there are no suitable methods to price the results after the reaction. It is not impossible for these procedures to be developed in the future and not so practical. DNA computers will be able to process related problems.
Dominik Heider, a professor of data science in Biomedicine at Philipps Marburg University, however, is somewhat skeptical about DNA-based computers: “From an academic point of view, it’s all quite different. But I fear it will continue to be irrelevant in practice.” He says the reason for this is quite easy; Everything that DNA computers can do, quantum computers can do. Heider says it’s also much easier to deal with them. “For videotapes, as in the days of VHS and Betamax, only one of the two approaches will work, and I doubt it will be DNA computers.”
Binary Data Translated into DNA
However, Heider is by no means willing to give up DNA for computer science. In the MOSLA Research Project, he is working with colleagues from computer science, biology, physics and chemistry to store data in DNA. To do this, binary data from conventional digital computers, that is, a long chain of zeros and ones, is translated into letters A, C, G, and T.
These letters represent the four bases. These letters mean the four bases that make up every DNA. Every genome of a living thing consists of a personal combination of these four bases. The translation can be easily transferred to real DNA, which can be stored for a long time in the laboratory and read again at any time. A digital computer can then convert the DNA data back into binary code and display the document digitally again.
However, Heider says there is a fair amount of computer science behind it in practice to ensure no data is lost on the way: “There are sources of defects in DNA storage during DNA synthesis, during replication. During storage and sorting, our mission is to develop fix codes that catch these errors.”
As with any storage, there is a trade-off between storage density and cost, he said: “Storing more information obviously costs more. Again, we can’t fit the code into an infinitely long segment of DNA. We need personal short modules, and then information on how to actually combine the modules again, always after sequencing.” But this meta information also takes up storage space.
Should Be Stored In A Cool, Dry And Dark Place
This DNA storage is still very valuable. “Until now, there was no need to produce DNA on such large scales,” Heider said. says. Therefore, a process that is inexpensive enough to make the use of DNA storage feasible in practice still does not exist. However, Heider hopes that in ten years, with enough research, things might look radically different.
For some applications, DNA storage has many unique advantages: “- Our technology will be used primarily for long-term archiving. Data like historical documents, birth records, or long-term weather data that is no longer changing is just great for DNA.” Once produced, storage requires almost no power except for the refrigerator to run. That’s because DNA is easy to hide.” Heider said that; cold, dry and dark”. Defect correction needs to compensate for individual mutations.
Most people will probably continue to store their vacation photos on hard drives, SSDs or the cloud. However, DNA storage may soon be the biological alternative for a large amount of information in archives that no one needs to have regular access to but is stored only for emergencies.